Genetic engineering
Engineering genomic neoantigens
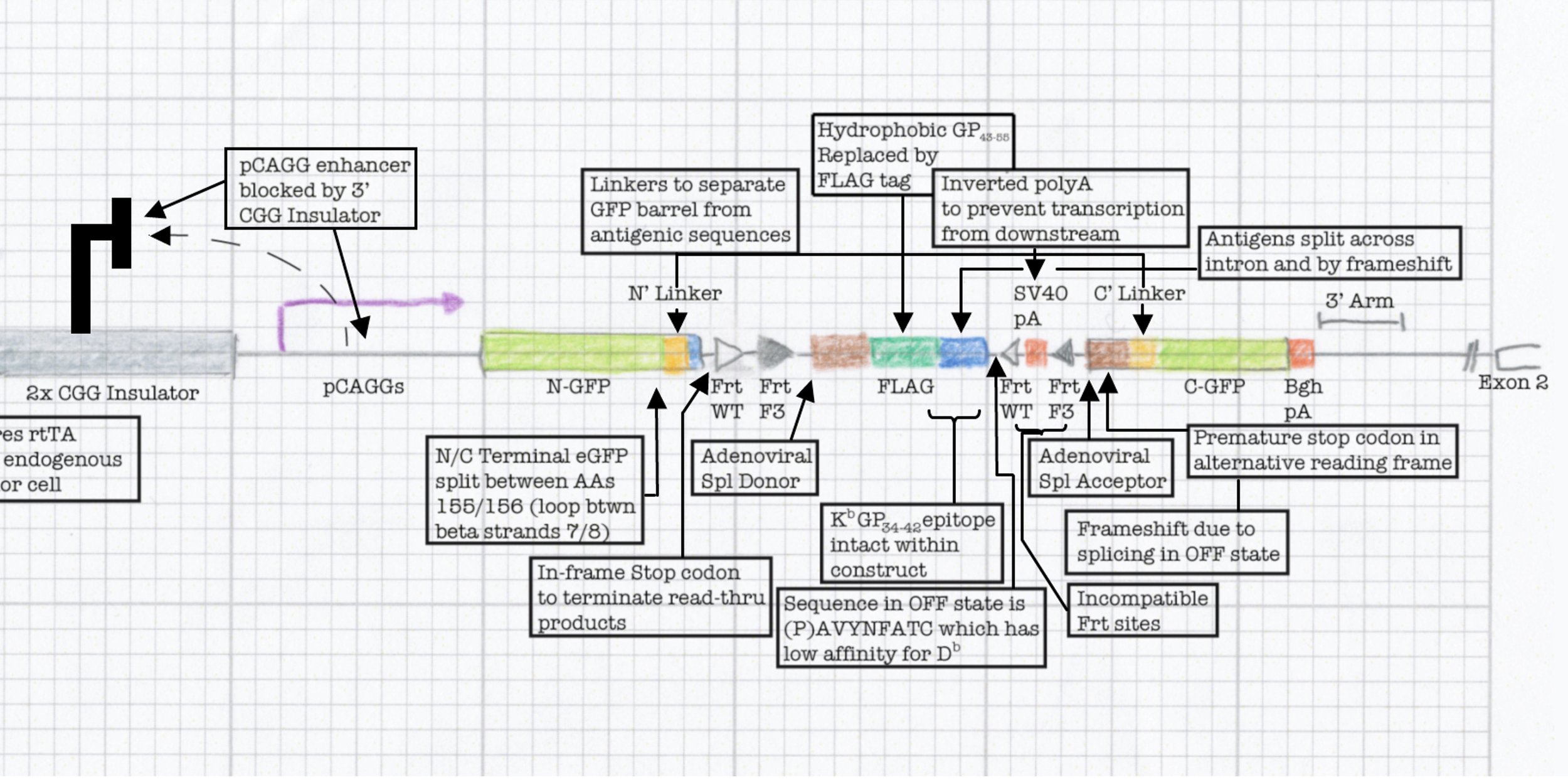
a Problem of leakiness
Developing thymocytes are very sensitive to the presence of their cognate antigen. As they transit through the medulla, thymocytes are exposed to many self antigens and the consequences of binding between the TCR and these self antigens can result in clonal deletion or development into a regulatory T cell fate (central tolerance). This process prevents autoimmunity, but is a challenge for the development of animal models where the goal is to have inducible neoantigens in the periphery. This is because any leakiness will result in central tolerance and it has traditionally been challenging to regulate antigen in a manner that prevents leaky expression in the thymus.
Engineering a neoantigen
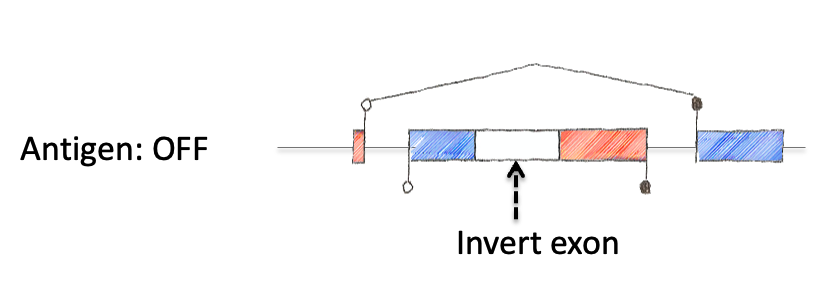
To tackle this engineering problem, we recognized that the neoantigen would need to be generated through the process of induction in the periphery. However, this would require both recombination based regulation and a means to encode the neoantigen in a manner that would produce proteins that were not affected by this machinery. Thus, we designed an neoantigen substrate with peptides that would be recognized by CD8 and CD4 T cells from LCMV (See concept figure). Key to this choice was the amino acid sequence KA, which allowed us to engineer splice donor/acceptor pairs into the DNA sequence, so that the transcribed RNA would splice out the intronic sequences.
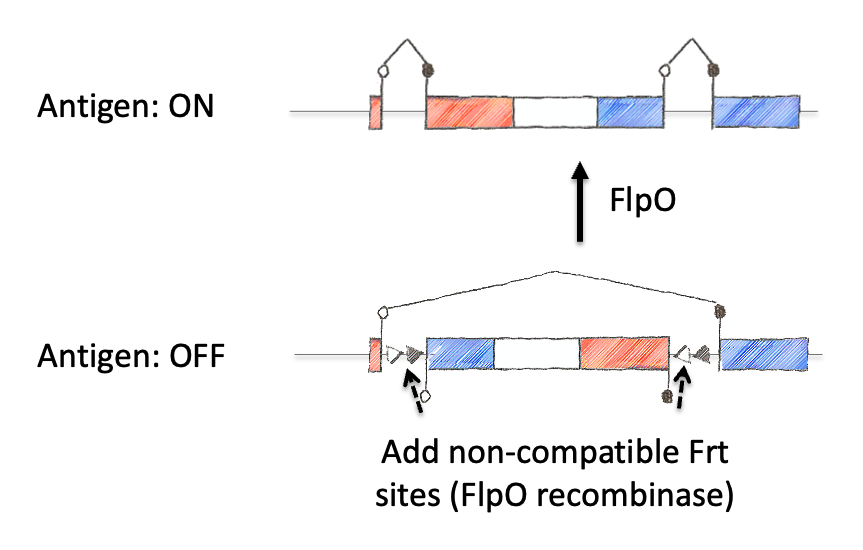
Next we inverted the central exon (exon 2) of the construct so that the antigenic sequences could not be produced, as they would be broken in half and inverted. As a means to induce expression, we needed to permanently invert exon 2. For this, we encoded machinery for recombination for the recombinase FLPo that consisted of two non-compatible Frt sites. This allowed a permanent inversion when the construct was exposed to FLPo protein, which would allow expression.
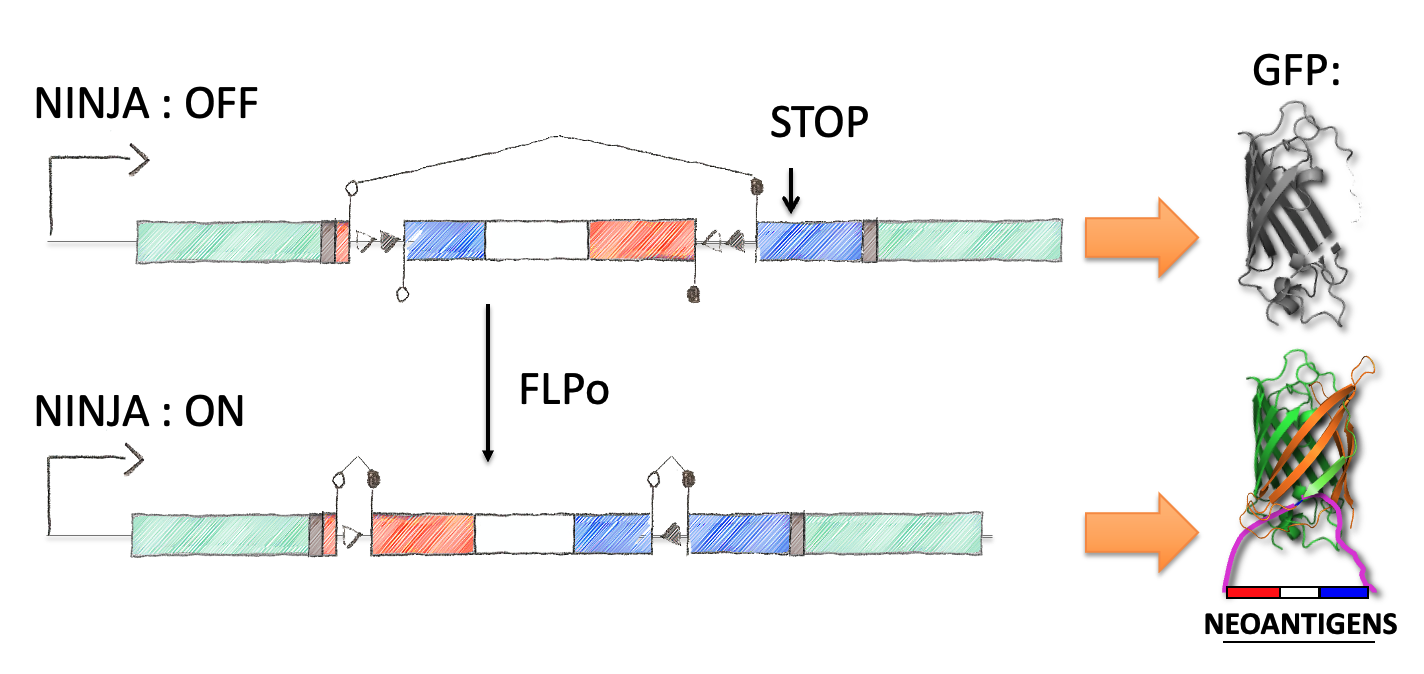
Next we focused on how to visualize that the neoantigen was expressed. We took advantage of work in the protein engineering field that has focused on modifying the fluorescent protein GFP to encode the neoantigen sequences in a loop of the GFP barrel. We also engineered the splicing so that direct splicing of exon 1 to 3 would be out of frame, and engineered a STOP codon in the alternative frame so that it would truncate the protein. This allowed for the OFF state to only produce half of the GFP protein (non-fluorescent) while the ON state would complement the reading frame and allow production of full length GFP that contained the neoantigen sequences. We extensively tested the neoantigen construct to make sure that the neoantigen was truly off in the OFF state and efficiently and faithfully transcribed and translated in the ON state.
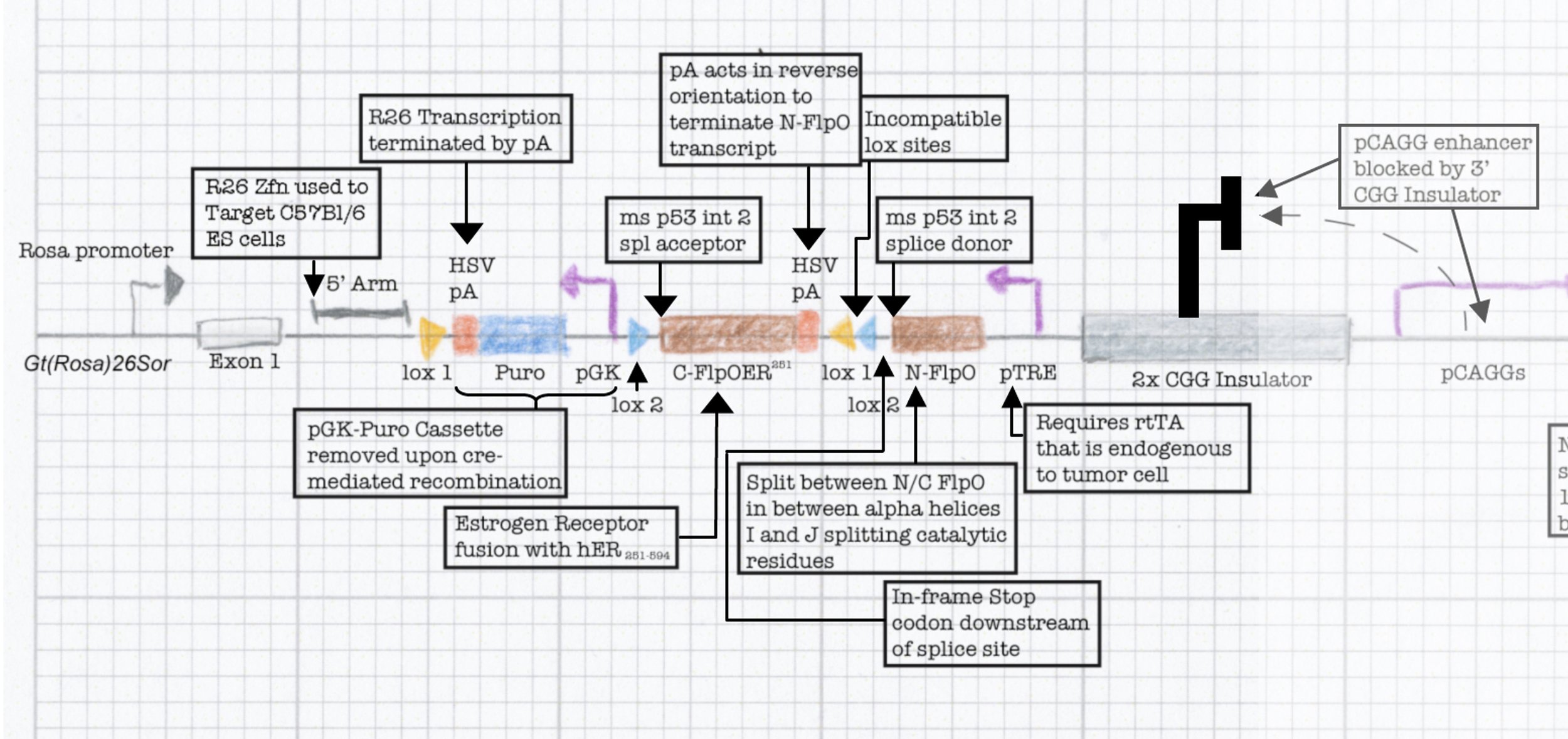
The NINJA model
This final construct was placed into a larger NINJA targeting and was known as the neoantigen module. NINJA also contained a “regulatory module” that allows for Cre/Doxycycline/Tamoxifen based regulation of FLPo activity. The result is an allele that requires Cre to become “poised”, and then simultaneous Dox/Tam treatment for induction. We further developed cell line models and the NINJA mouse, which has become the workhorse model for many of our studies of T cell immunology.
Ongoing work focuses on engineering advanced versions of the NINJA model to allow for better control over neoantigen expression and allow us to ask questions about how neoantigen encounter shapes both T cell immunobiology and cancer biology.
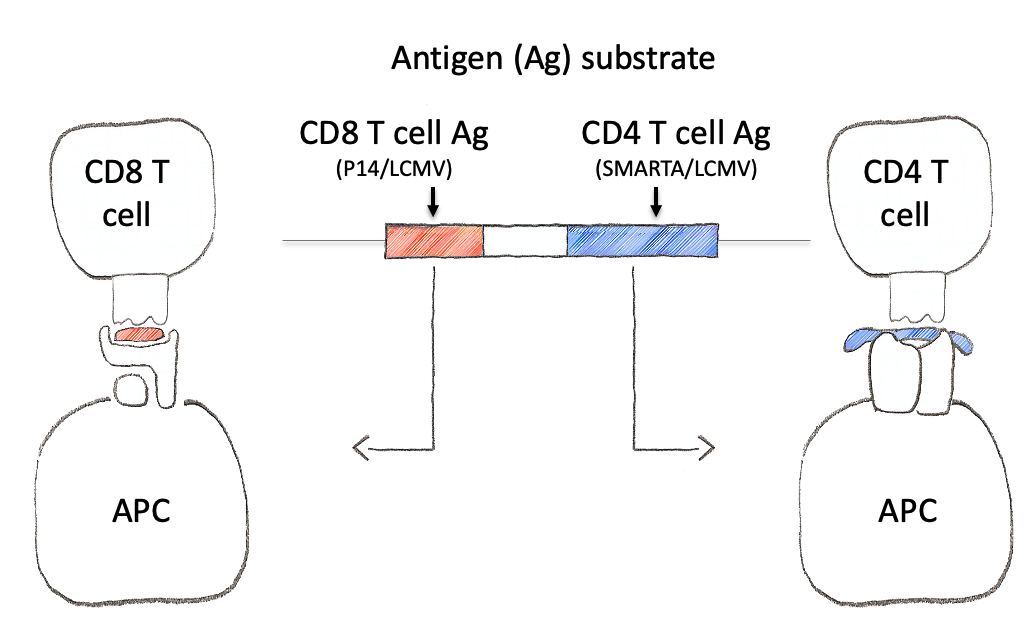
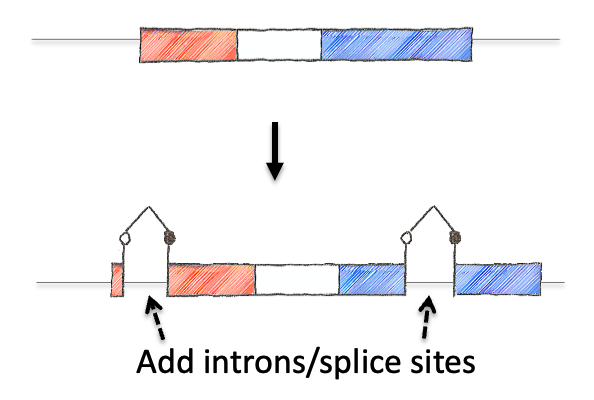
The concept of the NINJA neoantigen module